2. CO2/N2 Unary Example¶
In this example, we fit temperature-dependent unary data from [PXSL14].
2.1. Initialization¶
We first load the necessary packages
>>> import pyomo.environ as pyo
>>> import matplotlib.pyplot as plt
>>> import pandas as pd
>>> from isotherm_models.unaryisotherm import LangmuirUnary
2.2. CO2¶
We first get the data from the data file
>>> data = pd.read_csv('data_sets/CO2_BEA.csv')
Using pandas, we can easily take a peek at the data we have input from our .csv file
>>> data.head()
P [atm] Q [mmol/g] T [K] adsorbate
0 0.029814 0.096491 273.0 CO2
1 0.055856 0.175439 273.0 CO2
2 0.109177 0.328947 273.0 CO2
3 0.246821 0.719298 273.0 CO2
4 0.313781 0.903509 273.0 CO2
Before solving the model, we convert the partial pressures to si units
>>> P_i = data['P [atm]']*101325 # convert to Pa -- si units
so that we can create the model
>>> co2_model = LangmuirUnary(P_i, data['Q [mmol/g]'], data['T [K]'], name='CO2')
and solve it
>>> co2_model.solve()
We then take a look at the results
>>> co2_model.get_R2_pyomo()
0.99796
>>> co2_model.get_objective()
0.007229102
>>> co2_model.dH_i.display()
dH_i : Size=1
Key : Value
None : -20780.90809523844
>>> co2_model.q_mi.display()
q_mi : Size=1
Key : Value
None : 8.95582798469325
>>> co2_model.k_i_inf.display()
k_i_inf : Size=1
Key : Value
None : 3.8656918601559114e-10
And save the results to a file
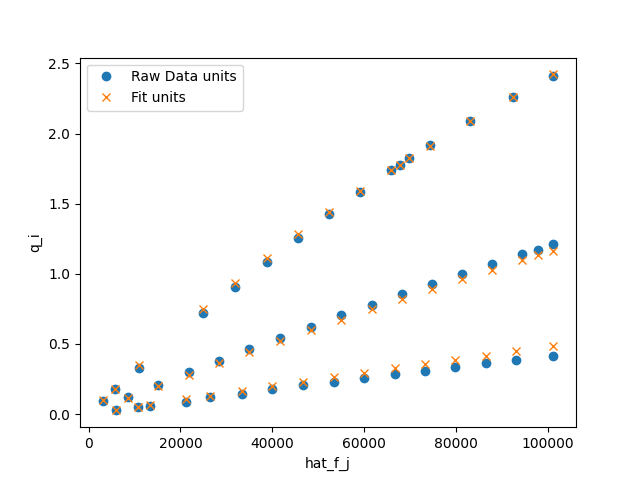
>>> fig = plt.figure()
>>> fig, ax = co2_model.plot_unary(fig=fig)
>>> _ = ax.legend()
>>> fig.savefig('docs/source/CO2_example.png')
which looks like
2.3. N2¶
We repeat a similar approach for the N2 isotherms, first formatting the data for input to the model
>>> data = pd.read_csv('data_sets/N2_BEA.csv')
Using pandas, we can easily take a peek at the data we have input from our .csv file
>>> data.head()
P [atm] Q [mmol/g] T [K] adsorbate
0 0.525470 0.070175 303.0 N2
1 0.592387 0.078947 303.0 N2
2 0.656824 0.083333 303.0 N2
3 0.722502 0.092105 303.0 N2
4 0.788179 0.100877 303.0 N2
Before solving the model, we convert the partial pressures to si units
>>> P_i = data['P [atm]']*101325 # convert to Pa -- si units
Instantiating (creating) the model
>>> n2_model = LangmuirUnary(P_i, data['Q [mmol/g]'], data['T [K]'], name='N2')
Solving it
>>> n2_model.solve()
We then take a look at the results
>>> n2_model.get_R2_pyomo()
0.99262
>>> n2_model.get_objective()
0.00194249
>>> n2_model.dH_i.display()
dH_i : Size=1
Key : Value
None : -12557.526993112784
>>> n2_model.q_mi.display()
q_mi : Size=1
Key : Value
None : 0.45280441671269905
>>> n2_model.k_i_inf.display()
k_i_inf : Size=1
Key : Value
None : 2.412336128388879e-08
And save the results to a file
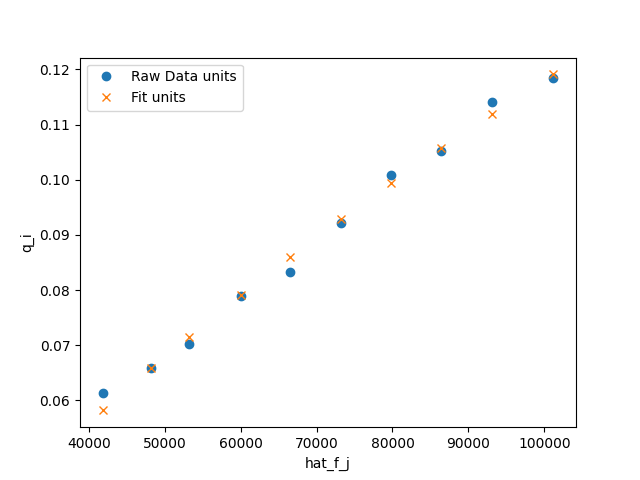
>>> fig = plt.figure()
>>> fig, ax = n2_model.plot_unary(fig=fig)
>>> _ = ax.legend()
>>> fig.savefig('docs/source/N2_example.png')
which looks like
2.4. Comparison to scipy¶
>>> import numpy as np
>>> popt, pcov = co2_model.solve_scipy()
>>> popt
array([ 3.71939309, -10.14817759, -7.28715595])
>>> popt - np.array(list(map(pyo.value, [co2_model.q_mi_star, co2_model.A_i, co2_model.H_i_star])))
array([3.28355311e-05, 3.67969745e-05, 3.88858220e-05])